The machine itself doesn’t look very impressive. In fact, it looks more like a home experiment…something you might use to distil your own gin or vodka.
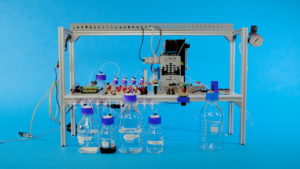
There are a few working parts: some glass bottles…some chemicals…a few sequencing devices. In total it cost researchers at Microsoft and the University of Washington about $10000 to make this prototype.
But this machine has huge potential. What we see here is a new type of device: one that will soon be capable of storing billions of times more data than your personal computer.
How? It’s all down to the material the machine uses to process information: DNA.
Consider its properties…
- You can fit 10 trillion molecules of this material into 1 cubic centimetre.
- With that small amount the computer would be able to perform 10 trillion calculations at a time.
- Just one pound of this material has the capacity to store more information that all electronic computers ever built.
Our DNA contains the collected information from millions years of evolution. All the instructions that have ever been passed between cells organisms bacteria and verterbrates are stored in DNA. DNA is basically nature’s storage system.
And its very stable. As Microsoft points out some DNA has held up for tens of thousands of years in mammoth tusks and the bones of early humans.
DNA is ideal for computer storage too. It’s extremely dense: compact enough to fit the entire internet in a shoebox. It’s easy to find. The energy required for storage is extremely low (less than a light bulb’s worth of power per year). And unlike the toxic materials use to make traditional microprocessors DNA biochips can be made cleanly.
In the past few years researchers have encoded all kinds of things in DNA strings mostly by hand: War and Peace, Deep Purple’s “Smoke on the Water”, a list of all the plant material archived in the Svalbard Seed Vault and a music video. Harvard researchers even created a living library by using E.Coli. In a paper published in Nature the researchers described how they used a Crispr system to insert bits of DNA encoded with photos and a GIF of a galloping horse into live bacteria.

When the scientists retrieved and reconstructed the images by sequencing the bacterial genomes they got back the same images they put in with about 90 percent accuracy.
How we build a DNA computer
But what was once done by hand can now be done by a machine. The prototype developed by Microsoft could speed up this process immeasurably.
In the demonstration, the machine performed a simple instruction…it converted the word “HELLO” into DNA and then back again.
There were a few steps.
- The device encoded the bits (1s and Os) into DNA sequences (A’s C’s T’s G’s).
- It then synthesized the DNA and stored it as a liquid.
- Next the stored DNA was read by a DNA sequencer
- Finally the decoding software translated the sequences back into bits.
In all the translation from bits to DNA and back again took 21 hours.
Researchers at Microsoft say that they have already found a way to reduce the time required by 10 to 12 hours.
So in order to replace existing silicon-chip storage technologies DNA will have to get a lot cheaper to predictably read, write and package.
It will take years to build a practical DNA computer. But that will happen….and when it does we might have computers that are extremely powerful.
Studying DNA computers may also lead us to a better understanding of complex systems that are still beyond our understanding – such as the microbiome and the human brain. In fact Timothy Lu’s team at Harvard has already been designing bacteria to produce working electronic circuit boards.
Lu’s group re-engineered bacteria DNA so that proteins would bind metals. This enabled them to program a pattern into biofilm – a bit like imprinting a pattern on fabric. Then they sprinkled gold atoms onto the biofilm to create pathways of gold wires. To complete the circuit board the scientists equipped the fibre with nanoscale semiconductors that emit light.
With new chips new circuit boards and sequencing devices – all based on biological material – we are entering a strange and very exciting new phase for computing.
Before then we’ll need to rely on chips that make use of silicon hybrids, photonics and neuromorphic computing. Drop us a line to discuss the companies involved in these areas: eoin@signumintel.co.uk
